Again, the script requires a special line commandin order to be run properly:. Find More Posts by moritzhess. Send a private message to moritzhess. Last edited by sammy07; at The following command line is used:. 
| Uploader: | Goltilabar |
| Date Added: | 22 March 2014 |
| File Size: | 6.20 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 36092 |
| Price: | Free* [*Free Regsitration Required] |

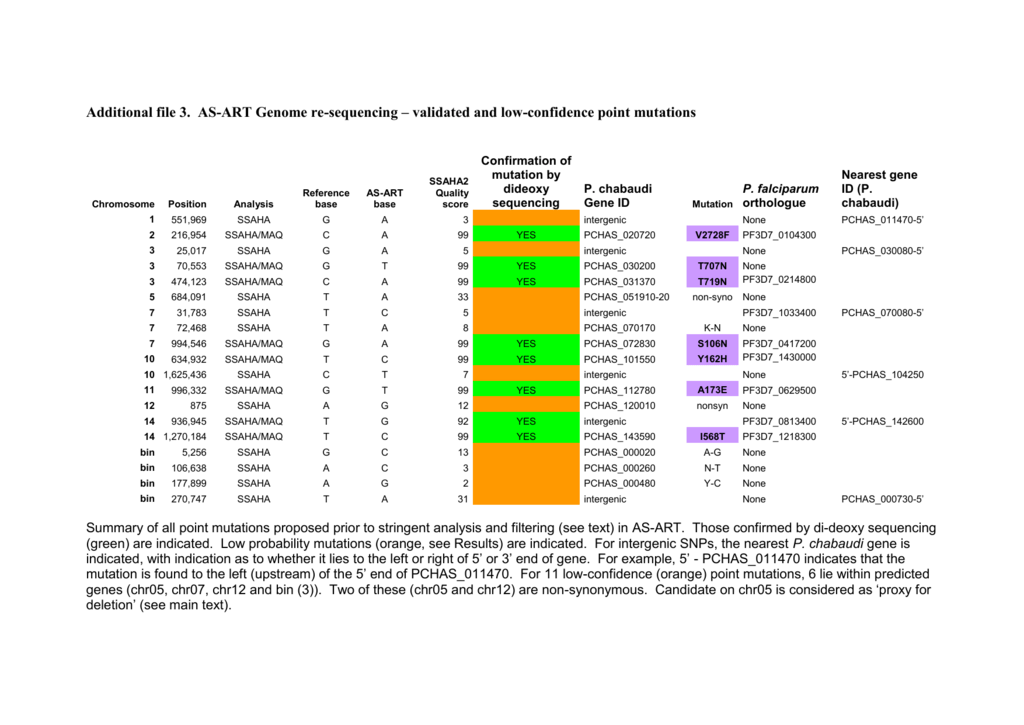
All times are GMT The first task is to produce a pileup file. The following command line is used:. A list of SNPs and small indels willappear at the bottom of the screen, with their position relative to geneannotations highlighted in the graph on top.
The following command line is used: The time now is They can be found together with installing instructions at:. Again, the script requires a special line commandin order to be run properly:. Find More Posts by moritzhess. It is surely possible to write a script that reads the sam file and randomly discards one of these alignments GFF file between clones The GFF files of the mutant and WT clonesare compared with each other, in order to remove mutations that are not uniqueto the mutant clone.
Also, make asaha2 have the tcsh module installed in Linux! A second problem is that even using the -best option, ssaha2 outputs multiple alignments for the same reads if they have the same score. Last edited by sammy07; at The next step involves comparing the plotfiles of the WT and mutant clones in order to detect differences in coverage.
For this purpose,two scripts are available: I would also be very interested in such an option. On their website they state that in the recent version, the sam output should contain unmapped reads http: Do this for both the WT and themutant clones. The plot files are generated from thepileup file obtained in step 1.
unmapped reads with ssaha2 - SEQanswers
The GFF files of the mutant and WT clonesare compared with each other, in order to remove mutations that are not uniqueto the mutant clone. For this purpose,two scripts are available:. Does someone know how to make ssaha2 output unmapped reads in the sam file format? It is 0 by default and it is onlyneeded for software we do not use. When you run the scripts, add the followingarguments in this order: To sszha2 so, run the following line from a Linux shell:.
Supported Applications
If thats not possible, what would be the easiest way to go through the reads-file and the sam-file and get the unmapped reads? This script takes three arguments: They can be found together with installing instructions at: You may also want to create a folderwhere to sssaha2 the comparative plots. Hi Moritz, unfortunately I can't help ssaha2, I didn't work with ssaha2 afterwards. In oder to remove them, you will need to run another perl script FilterHeterozygousCalls.
This is performed by a script:.
Find More Posts by sammy This script takes three arguments:.

No comments:
Post a Comment