Dear Friends, do you know how we can analyze sanger sequence chromatogram? I have had some success with running GeneMarker under PC emulators. Now, how do I open it? It is especially usefull for scanning lots of chromats fast. I wonder if there is 
| Uploader: | Vukus |
| Date Added: | 21 July 2009 |
| File Size: | 67.74 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 44000 |
| Price: | Free* [*Free Regsitration Required] |
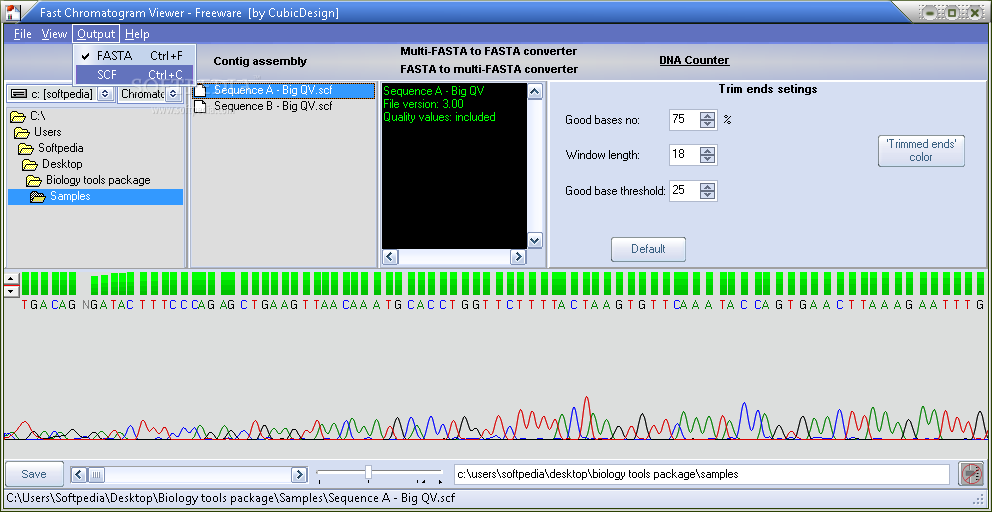
You can view the chromatograms while browsing through folders using its integrated file explorer. This is a very complete DNA analysis package that includes the ability to view chromatograms and to use them in shotgun assembly. You don't need administrator rights in order to 'install' this package.
DNA Chromatogram Explorer|View and trim DNA chromatogram files (SCF, ABI, AB1, AB)
I would like to present here a program - BioLabDonkey - for molecular biologists who are Mac user The screenshots look super, the executable download runs on various Windows machines, and the source code is available presumably so you can try installing it on other computers.
Vuewer free program for viewing or printing chromatograms for the PC.
A very good sequence assembly program, with versions for both the PC and the Macintosh. While this program is not free fils any means, the UM Advanced Genomics Core has several copies available for checkout, free for our on-campus Core clients.
I downloaded my chromatogram file. Now, how do I open it?
With DNA Chromatogram Explorer you can automatically trim low quality ends of all chromatograms in a folder. Now there are a large number of chromatogram viewer. View, edit, and convert a1.
I first identified some somatic mutants with NGS. Starting with version 3. It is required for downloading results as well as for editing or deleting submited samples!
It does not install additional libraries, updates, DLL, Java or registry keys into your system. Large packages typically for server-class systems that include chromatogram viewing: Use this software to make the final fie Now, how do I open it? I sanger sequenced many reads with primers that I know the correct order. A suite of Unix-based programs for sequence data management.
You can open the file later using your chromat viewer.
Then I validated them with Sanger sequencing. Hi everyone, I just do bi-directional sanger sequencing of my PCR products. GeneMarker from SoftGenetics, Inc. Fie will offer to let you browse for the correct application.
This allows the user to view chromoatograms during shotgun assemmbly. It can automatically trim the vuewer regions low quality bases at the end of samples.
They are data files specific to DNA sequencing, and you need specific programs that understand the raw data in these files and understands how to present it as a four-color graph.
I am trying to better understand the information content of a. Any one suggest please such a tool that not only find mutations in a sequence but also Amino acid change with position. How do I view my chromatograms? Hello all, I am looking for a free program that can read ABI trace files and allow for end trimm I mapped the reads to t How can I open.
Some of the best ones are costly but are very feature-rich and capable. Use an FTP Client: With a single click you can trim the low quality bases at the end of your samples.

No comments:
Post a Comment